Introduction
The overuse of carbapenem antibiotics has resulted in an increase in carbapenem-resistant gram-negative bacterial infections, posing a significant public health threat. These infections are prevalent in intensive care units (ICUs) due to invasive procedures, low immunity, and illness severity. Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa are the most common organisms responsible for infections. Due to the strong resistance of these bacteria, treatment options are limited. Predictive models for CR-GNB infections in ICU patients have produced inconsistent results, so it is crucial to identify risk factors and create a reliable predictive model for individualized treatments. The study retrospectively analyzed data from ICU patients to provide a scientific basis for clinical treatment and prevention measures for CR-GNB infections.
Patients and methods
Pathogenic bacteria and antibiotic
susceptibility testing
Identification and routine antibiotic susceptibility testing of pathogenic bacteria were performed using the Vitek 2 Compact automated identification or antibiotic susceptibility testing system from bioMerieux (Lyon, France). Antibiotic susceptibility test results were interpreted using the 2017 edition of the Performance Standards for Antimicrobial Susceptibility Testing published by the Clinical and Laboratory Standards Institute.
Data Collection
The collected data from patient medical records includes age, sex, underlying diseases, history of recent hospitalization, antibiotic use, and several other parameters such as SOFA and APACHE II scores, white blood cell count, and pathogenic bacteria.
Statistical analysis
The data was analyzed using Stata SE15, with non-normally distributed variables compared using Mann-Whitney U test and categorical data compared using X2 test. Independent risk factors for CR-GNB infection were determined using multivariate logistic regression analysis, and a nomogram-based predictive model was constructed. The model’s stability and predictive ability were evaluated using the Hosmer-Lemeshow goodness-of-fit test and the area under the receiver operating characteristic curve (AUC). The model’s practical value was determined using decision curve analysis. Statistical significance was set at p < 0.05.
Results
Distribution of pathogenic bacteria
and sites of infection
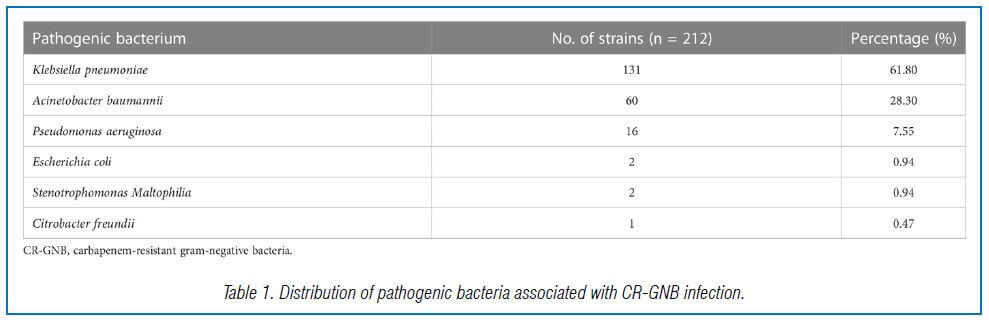
This study included 309 ICU patients with GNB infection out of 908 admitted between December 2017 and September 2020. Of these, 212 patients had CR-GNB infection, with carbapenem-resistant K. pneumoniae being the most common strain, followed by carbapenem- resistant A. baumannii and carbapenem-resistant P. aeruginosa (Table 1).
Antibiotic susceptibility test results of
CR-GNB main types
The study found that CRKP strains had low resistance to tigecycline and polymyxin, but high resistance rates to all other commonly used antibiotics. CRAB showed high susceptibility to tigecycline and polymyxin, but high resistance rates to ticarcillin-clavulanic acid, cefoperazone-tazobactam, piperacillin-tazobactam, ceftazidime, ceftriaxone, cefepime, aztreonam, ciprofloxacin, and levofloxacin. CRPA strains showed good in vitro anti-bacterial activity against piperacillin-tazobactam, ceftazidime, cefepime, amikacin, and polymyxin, but high resistance rates to tobramycin, ciprofloxacin, levofloxacin, aztreonam, and trimethoprim-sulfamethoxazole.
Comparison between the groups of
patients in the experimental cohort
205 ICU patients with GNB infection were studied. Of them, 62 had CS-GNB infection, and 143 had CR-GNB infection. Patients with CR-GNB infection had more recent hospitalization, carbapenem use, central venous catheterization, and combination antibiotic treatment, and higher incidence of septic shock and in-hospital mortality than patients with CS-GNB infection. No significant difference was observed in sex, age, and other factors between the two groups.
Nomogram-based predictive model of
CR-GNB infection in ICU patients
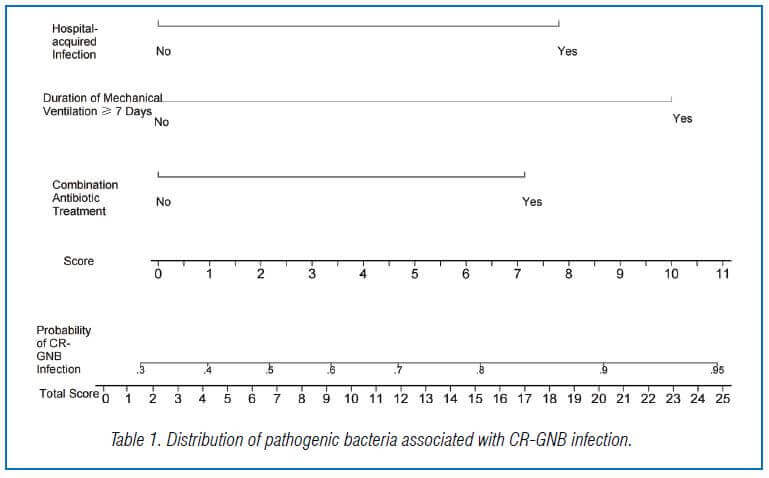
A nomogram-based predictive model for CR-GNB infection in ICU patients was created using independent risk factors identified from multivariate logistic regression analysis. The formula for the predictive model was P = 1/[1+exp [-(-1.090 + 1.162X1 + 1.271X2 + 1.628X3)]]. The scores of various independent variables were used to calculate an overall score of 0-24.9 points, with combination antibiotic treatment, hospital-acquired infection, and duration of mechanical ventilation ≥ 7 days having scores of 7.1, 7.8, and 10.0 points, respectively. The nomogram showed that a total score >16 points predicted a probability of >80% for the occurrence of CR-GNB in ICU patients (Figure 1).
Internal validation of the predictive model
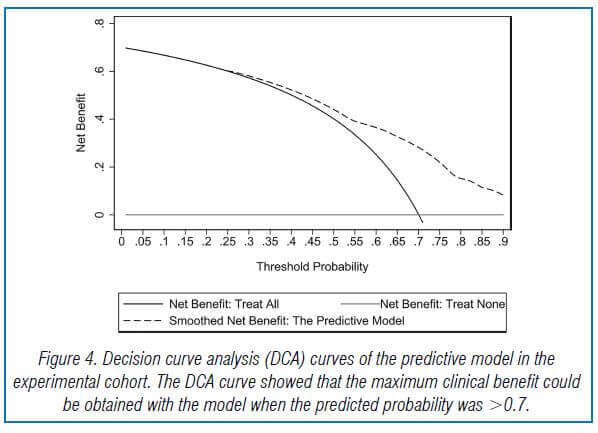
The model’s goodness of fit was assessed using the Hosmer-Lemeshow test, and it showed a good fit with the observed data. The AUC of the predictive model was 0.753, indicating good predictive performance and discriminatory power. The model’s repeatability was also good. The DCA curve showed that the model could provide high practical value in clinical practice when the predicted probability was >0.7.


Validation of the proposed predictive
model
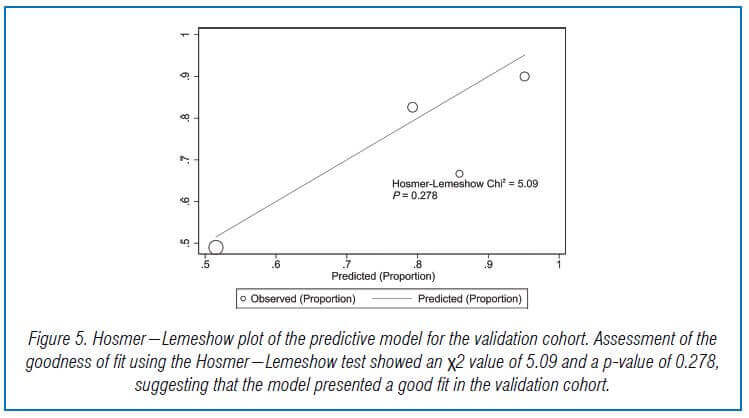
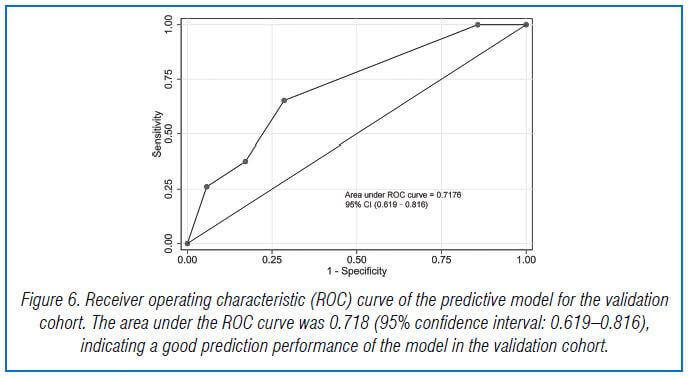
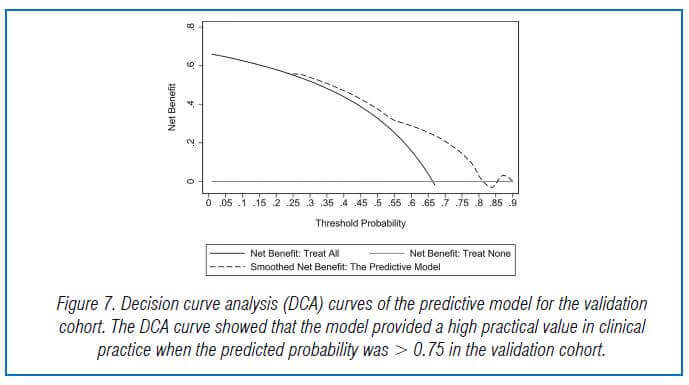
In the validation cohort, there were 104 GNB infection patients, of which 35 had CS-GNB infection and 69 had CR-GNB infection. The Hosmer-Lemeshow test showed a good fit of the model, with a p-value of 0.278. The AUC was 0.718, indicating good prediction performance in the validation cohort, and the DCA curve showed high practical value when the predicted probability was >0.75 (Fig 5,6,7).
Conclusion
In summary, this study investigated antibiotic resistance in CR-GNB and identified CRKP, CRAB, and CRPA as the most common in ICU patients. Using independent factors, a nomogram for predicting the risk of CR-GNB infection in ICU patients was developed and validated. These findings could aid in personalized treatment and the implementation of effective control measures to reduce the risk of CR-GNB infection, particularly in ICU settings.
Source: Liao, Qiuxia, et al. “Carbapenem-resistant gram-negative bacterial infection in intensive care unit patients: Antibiotic resistance analysis and predictive model development.” Frontiers in Cellular and Infection Microbiology 13 (2023): 65.