Introduction
Infection is one of the most common complications for patients with neutropenia and can lead to high mortality if inappropriate empirical antibiotic treatment is administered. Accurate and timely pathogen detection is critical in optimizing antibiotic use; however, it is challenging in patients with neutropenia because of nonspecific clinical symptoms, signs, and the low positivity rate of conventional microbiological tests (CMT).
In recent years, microbial next-generation sequencing (NGS) has emerged as a promising method for pathogen detection in patients with neutropenia. NGS is a fast, non-culture-based, and unbiased method that has garnered considerable scientific attention in the field of etiological diagnosis. However, invasive specimens such as bronchoalveolar lavage fluid (BALF) and tissues are often required for NGS, which can lead to complications such as hypoxemia and endobronchial bleeding in immunocompromised patients.
Plasma cell-free DNA (cfDNA) NGS, a non-invasive microbial identification technology, has shown great potential in the diagnosis of clinical infectious diseases such as bloodstream infection (BSI), invasive fungal infection, endocarditis, sepsis, and infection associated with chimeric antigen receptor-T. However, only a few studies have focused on the application of plasma cfDNA NGS in patients with hematological diseases, and the clinical impact of cfDNA NGS is unclear.
Therefore, the article aims to assess the pathogen diagnostic ability and clinical impacts of plasma cfDNA NGS in patients with hematological diseases, particularly those with neutropenia. The study has significant implications for the management of patients with neutropenia, as timely and accurate pathogen detection can help optimize antibiotic use and improve patient outcomes.
Methods
This study retrospectively analyzed the results of plasma cell-free DNA sequencing performed on 184 and 163 specimens collected from hematological patients suspected of infections with (Group I) or without (Group II) neutropenia, respectively. The records of patients who undertook the plasma cfDNA NGS test for suspected infections at the Institute of Hematology and Blood Diseases Hospital were reviewed. (Fig 1)

The diagnostic performance and the clinical impact of plasma sequencing were comparatively evaluated to conventional microbiological tests and a composite reference standard (conventional tests combined with the clinical assessment). We retrospectively reviewed the records of patients who undertook the plasma cfDNA NGS test for suspected infections at the Institute of Hematology and Blood Diseases Hospital.
Results
The overall positive detection rate of plasma cell-free DNA sequencing was significantly higher than that of conventional microbiological tests (72.6% vs.31.4%, P < 0.001). The positive rate of conventional microbiological tests in Group I was lower than that in Group II (25.5% vs. 38.0%, P = 0.012).
Combining plasma sequencing with conventional tests yielded a positive detection rate of 75.0% and 74.8% for these two groups, respectively. Using the composite reference standard, the sensitivity and specificity of plasma sequencing were 89.1% and 65.1%, respectively.
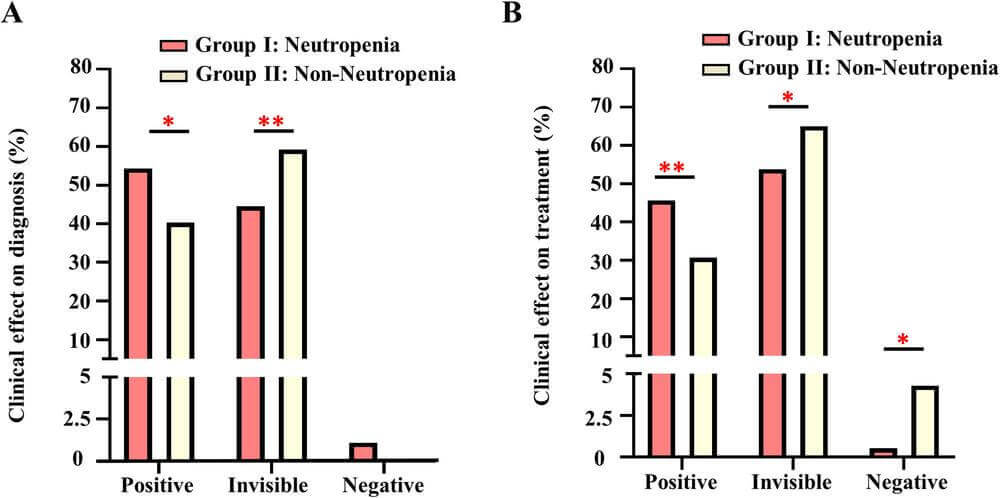
The proportions of the positive impact of cell-free DNA sequencing results in the Group I were higher than in the Group II in terms of both diagnosis and treatment (diagnosis: 54.3% vs. 40.5%, P = 0.013; treatment: 45.7% vs.30.7%, P = 0.004). A total of 73 patients (21.0%) benefited from plasma sequencing through adjustment of the antibiotic regimen.
Conclusion
The diagnostic yield of conventional microbiological tests was low in patients with neutropenia. Combining conventional tests with plasma cell-free DNA sequencing significantly improved the detection rate for pathogens and optimized antibiotic treatment. Our findings on the clinical impact warrant confirmation through larger, multicenter, randomized controlled trials. Moreover, the cost-effectiveness of this testing strategy remains unknown and requires further exploration.
Source: Xu, Chunhui, et al. “Utility of plasma cell-free DNA next-generation sequencing for diagnosis of infectious diseases in patients with hematological disorders.” Journal of Infection 86.1 (2023): 14-23.




